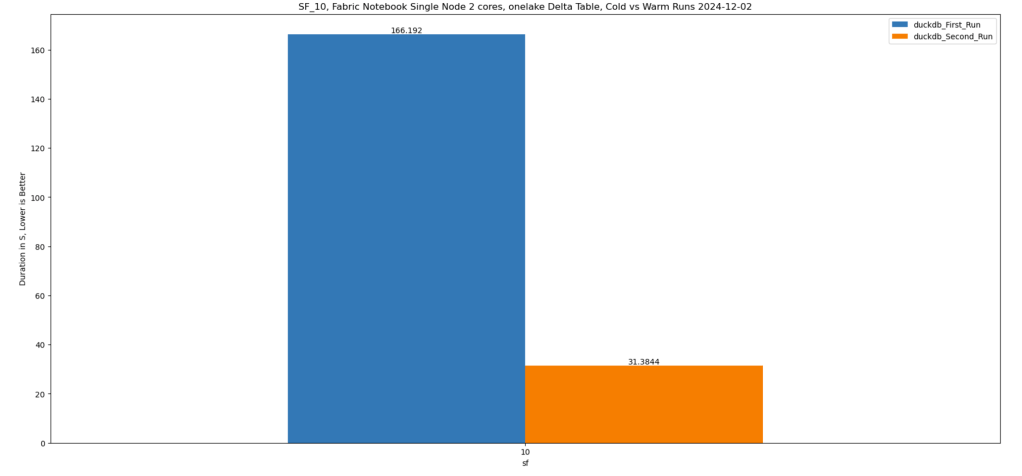
This weekend, I came up with an idea to speed up query execution when running DuckDB inside a Fabric Notebook—and it actually works! 🎉
You can download the notebook here
Approach
- Parse the Query
- Use SQGLot to parse the SQL query and extract the list of Delta tables that need to be scanned from OneLake.
- Track Table Metadata
- Capture the Delta table version and ID to ensure consistency.
- Selective Copy
- Copy only the necessary tables locally to satisfy the query.
- Reuse Cached Data
- For subsequent queries, check if the Delta table has changed:
- If unchanged, read data from the local SSD.
- If new tables are required, repeat the caching process for those tables.
- For subsequent queries, check if the Delta table has changed:
Why It Works
This approach effectively creates a temporary, ad hoc disk cache in the notebook. The cache:
- Persists only for the session’s duration.
- Evicts automatically when the session ends.
- Ensures consistency by validating whether the Delta table in OneLake has changed before reusing cached data.
- Thanks to the Delta format, this validation is a relatively cheap operation.
- Leverages the user-level isolation in Fabric notebooks to eliminate risks of data inconsistency.
Despite its simplicity, this method has proven to be highly effective for query acceleration! 🚀
Limitations
Yes, I know—the cache is rather naïve since it loads the entire table. Other systems go further by:
- Copying only the columns needed for the query.
- Fetching just the row groups relevant to the query.
However, these optimizations would need to be implemented natively by the engine itself.
Industry Gap
Although virtually all Python engines (e.g., Polars, DataFusion, etc.) support reading formats like Delta and Iceberg, almost none offer built-in disk or RAM caching. This lack of caching support limits performance optimization opportunities.
Hopefully, this will change in the future, enabling more efficient workflows out of the box.
Btw, this is really fast !!! just a hint, this is faster than the results obtained by a state of the art DWH in 2022 !!!